Single-cell lactate production rate as a measure of glycolysis in endothelial cells
Devin Harrison,1,2,3,4 David Wu,2,4,5 Jun Huang,1,3,* and Yun Fang1,2,6,**
1Graduate Program in Biophysical Sciences, The University of Chicago, Chicago, IL 60637, USA
2Department of Medicine, Biological Sciences Division, The University of Chicago, Chicago, IL 60637, USA
3Pritzker School of Molecular Engineering, The University of Chicago, Chicago, IL 60637, USA
4These authors contributed equally
5Technical contact
6Lead contact
*Correspondence: huangjun@uchicago.edu
**Correspondence: yfang1@medicine.bsd.uchicago.edu
Summary
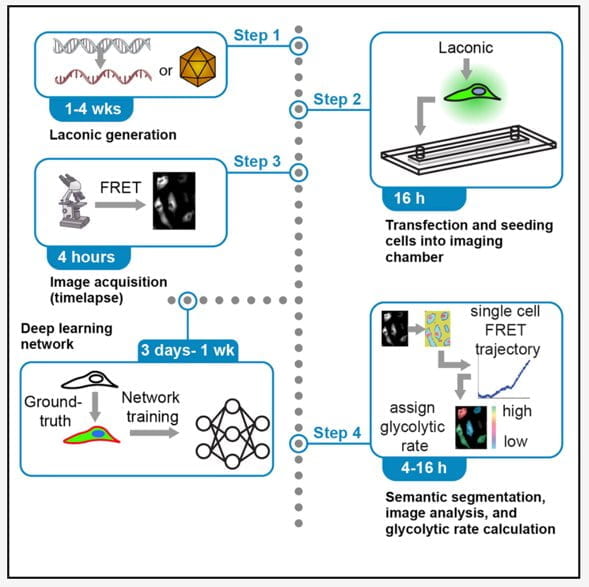
Heterogeneous metabolism supports critical single-cell functions. Here, we describe deep-learning-enabled image analyses of a genetically encoded lactate-sensing probe which can accurately quantify metabolite levels and glycolytic rates at the single-cell level. Multiple strategies and test data have been included to obviate possible obstacles including successful sensor expression and accurate segmentation. This protocol reliably discriminates between metabolically diverse subpopulations which can be used to directly link metabolism to functional phenotypes by integrating spatiotemporal information, genetic or pharmacological perturbations, and real-time metabolic states.